Abstract
Introduction: Myeloproliferative Neoplasms (MPN) are blood diseases caused by mutations activating JAK2 in hematopoietic stem cells (HSCs) which lead to clonal expansion and overproduction of myeloid lineages. MPN patients are at increased risk for transformation to bone marrow fibrosis (myelofibrosis) and acute myeloid leukemia (AML), both of which are associated with poor clinical outcomes. Importantly, targetable mechanisms to prevent progression remain elusive. The High Mobility Group A1 (HMGA1) gene encodes chromatin regulators which are enriched in hematopoietic stem cells and aberrantly overexpressed in MPN with highest levels after transformation to myelofibrosis and acute leukemia (Li & Kim et al Blood 2022, Xian et al Nature Commun 2017). We recently discovered that HMGA1 is required for progression to leukemia in murine models of JAK2V617F MPN leukemia. Further, loss of just a single Hmga1 allele prevents progression to myelofibrosis in JAK2V617F transgenic mice. Emerging evidence suggests that immune escape represents a fundamental pathway involved in progression in diverse tumors. We therefore sought to: 1) test the hypothesis that HMGA1 drives MPN progression by dysregulating gene networks involved in immune evasion, and, 2) identify mediators of immune escape that could be disrupted in therapy.
Methods: To identify gene networks regulated by HMGA1 in JAK2V617F MPN, we performed multi-omics sequencing analyses, including RNAseq, chromatin immunoprecipitation (ChIPseq), and assays of chromatin accessibility (ATACseq) from JAK2V617Fmutant AML cell lines (DAMI, SET-2) + HMGA1 depletion. HMGA1 gene expression was inactivated using CRISPR/Cas9 or short hairpin RNAs (shRNA) targeting 2 different sequences. We used gene set enrichment analysis to dissect molecular mechanisms underlying HMGA1. Transcriptional networks were validated at the level of mRNA and protein via quantitative RT-PCR and flow cytometry. To identify drugs to disrupt HMGA1 immune evasion networks, we applied the Broad Institute Connectivity Map (CMAP) and cytotoxicity assays. To validate these pathways in human MPN, we assessed gene expression in independent MPN patient cohorts.
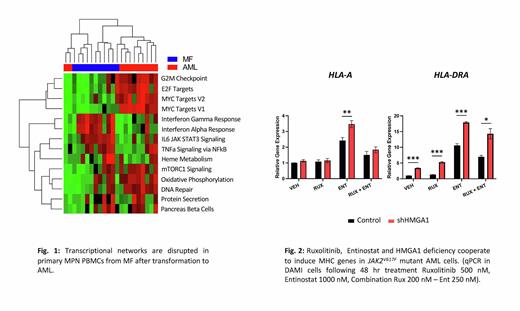
Results: To elucidate transcriptional networks governed by HMGA1 during MPN progression to leukemia, we integrated RNAseq, ChIPseq, and ATACseq in JAK2V617F mutant AML cells (DAMI, SET-2) which revealed that HMGA1 represses genes involved in antigen presentation (Interferon γ response networks), including genes encoding major histocompatibility complex (MHC) class I and II antigens. Silencing HMGA1 via CRISPR or shRNA results in up-regulation of MHC class I and II antigen genes, with greatest induction of MHC class II genes HLA-DRA/DRB1. Similarly, HMGA1 depletion increases cell surface expression of HLA-DR antigens by flow cytometry. Further, inhibiting HMGA1 in JAK2V617Fmutant AML cells results in up-regulating the class II major histocompatibility complex transactivator (CIITA) and invariant chain (CD74), which function in inducing MHC class II gene expression and chaperoning structure assembly, respectively. To identify underlying mechanisms, we intersected ChIPseq with ATACseq, which revealed HMGA1 occupancy downstream of the HLA-DRB1 locus, including regions at +6.27kb, +6.7kb, +14kb, and +15kb from the transcription start site. Further, preliminary results suggest that HMGA1 recruits repressive histone marks to repress HLA-DRB1 at these sites. To target HMGA1 networks, we queried CMAP, which identified the histone deacytelyase inhibitor (HDACi), entinostat. Strikingly, entinostat not only up-regulates MHC class II genes and antigens, but is also cytotoxic in JAK2V617F mutant AML cells. Finally, HMGA1 depletion enhances sensitivity to entinostat, which synergizes with JAK inhibitor, Ruxolitinib, to further induce MHC gene expression.
Conclusions: We discovered a novel epigenetic program whereby HMGA1 drives immune evasion during MPN progression by binding to chromatin and modulating transcriptional networks to repress MHC antigen presentation. Most importantly, HMGA1 immune evasion networks are dysregulated in human MPN after progression to AML. Together, our studies reveal a new paradigm whereby HMGA1 down-regulates MHC antigens during MPN progression, suggesting that targeting HMGA1 networks with entinostat could activate an immune attack and prevent MPN progression.
Disclosures
Rampal:Zentalis: Consultancy, Research Funding; Promedior: Consultancy; CTI: Consultancy; AbbVie: Consultancy, Membership on an entity's Board of Directors or advisory committees; PharmaEssentia: Consultancy; Galecto: Consultancy, Membership on an entity's Board of Directors or advisory committees; Sunimoto Dainippon: Consultancy; Disc Medicines: Consultancy; Jazz Pharmaceuticals: Consultancy; Stemline: Consultancy, Research Funding; Gilead: Consultancy; Sierra Oncology: Consultancy; Stemline: Consultancy, Research Funding; Blueprint: Consultancy; Kartos: Consultancy, Membership on an entity's Board of Directors or advisory committees; Novartis: Consultancy; Celgene/BMS: Consultancy; Incyte: Consultancy, Research Funding; Constellation Pharmaceuticals, Inc., a MorphoSys Company: Consultancy, Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal